MetaChrom
Last Updated: Oct 2020
This server is an implementation of the MetaChrom model, a deep transfer learning framework to predict regulatory effects of noncoding genetic variants at single-nucleotide resolution. MetaChrom leverages both an extensive compendium of publicly available epigenomic data and epigenomic profiles of cell types related to specific phenotypes of interest. This web server can be used to predict variant effects in a customized neurodevelopment model (please see the paper for details).
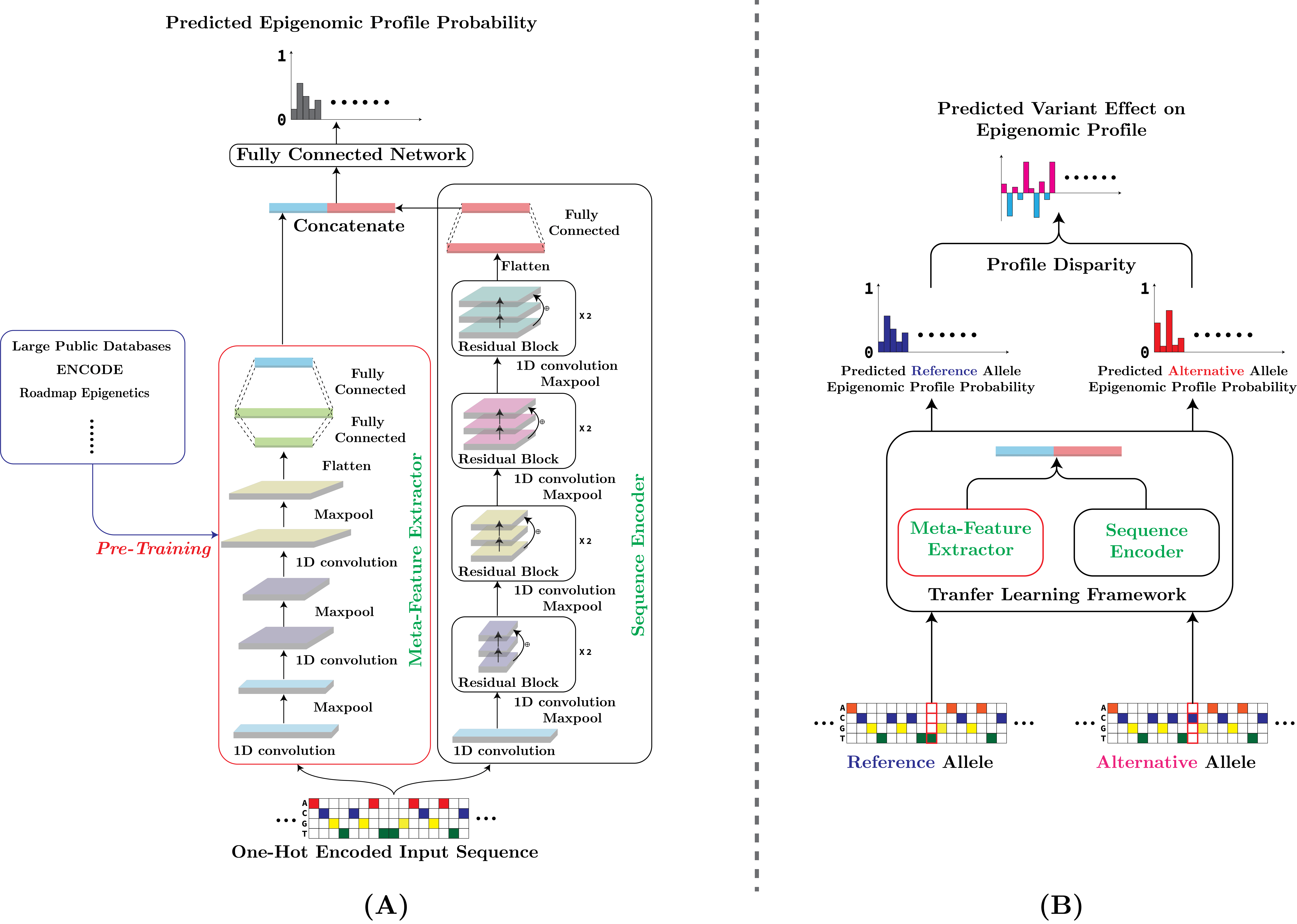
- Overall architecture of MetaChrom. The input sequence is fed into both the meta-feature extractor and the ResNet sequence encoder. Their outputs are then concatenated for the prediction of epigenomic profiles.
- Pipeline for predicting variant effects on sequence epigenomic profile.
Submit a file for analysis
Only supports GRCh38/hg38 genome coordinates!

